Challenge
To take a step further in personalized cancer medicine by integrating clinical, genomic, anatomopathological and radiomic data and use AI models to help choose the most appropriate treatment.
Solution
A multimodal platform to store, process and integrate all patient datasets in a single centralized repository for AI modeling, with a focus on continuous and iterative model training.
Main goal
The project is expected to provide objective guidance for individual clinical management but also to help develop national health strategies to prevent, early diagnose and treat the most prevalent tumors in a more cost-effective way.
About DIPCAN
DIPCAN is the largest publicly funded project in Spain for an artificial intelligence (AI) oncology platform. The project’s goal is to revolutionize clinical practice in oncology by utilizing personalized medicine and integrating various data sources – that is clinical, radiomic, anatomopathological, and genetic – from 2,000 patients with metastatic cancer. Genomcore is providing its platform within the project to process, standardize, and integrate the data sources into a single database for its later use in AI models.
The project has the presence of Eurofins Megalab, MD Anderson International Foundation Spain and five SMEs, and counts with €7,5M in funding from the European Union through the Spanish Ministry of Economic Affairs and Digital Transformation.
- Spain
- 7 leading entities
- https://estudiodipcan.com/
The study is based on the digitalization and management of personalized medicine in oncology. The goal is to include a total of 2,000 patients older than 18 years from all over Spain with non-hematological metastatic solid tumors. The study is purely observational meaning that no treatment that would enhance the survival or quality of life of the patients will be provided. During the study, a set of diagnostic tests will be performed – analysis, magnetic resonance imaging, pathological anatomy and molecular and tumor expression analysis – and the results of all these tests will be integrated with the aim to create an AI algorithm to predict treatment response. In gratitude for their valuable contribution, patients will have access to specific diagnostic tests that are otherwise very hard to access, like the molecular study, and will receive for free all the obtained results in an easily understandable report.
“Digital transformation must reach the field of healthcare and what better field than oncology to lead this transition. [...] The team of the project believes in personalized medicine and, if the algorithm that has been designed provides the expected results, it would identify some patients that could be more likely to better respond to each type of treatment when personalized ”
Dr. Enrique Grande, the Principal Investigator and Head of the Medical Oncology Service and Clinical Research at the MD Anderson Cancer Center Madrid Foundation.
CHALLENGE
Multidisciplinary and integrated care for cancer patients is more crucial than ever. Instead of a unidimensional, genomic-focused approach, we are transitioning towards a more comprehensive and holistic view of the tumor microenvironment. Each type of omic data provides a list of molecular differences related to cancer, revealing useful markers of the disease and insights into biological pathways. However, individual omics data strands alone are insufficient to determine the causal relationship between molecular signatures and the development of cancer. By integrating different types of biomedical data, it is possible to first, identify these causative changes and second, improve our understanding of the variability in treatment response, two major challenges in oncology. However, due to the intricate nature of omics and precision medicine, the integration and standardization of heterogeneous data types is a complex but also crucial step. Genomcore participates in the DIPCAN study as the leader in the governance, processing and integration of multi-omics data.
SOLUTION
Genomcore’s platform offers a GDPR and HIPAA-compliant framework for the store, management, standardization and integration of all kinds of datasets. This centralized data storage and analysis environment in the cloud is accessible from anywhere and can be adapted to the unique needs of each project so that participants can share data, metadata, analysis tools and results, facilitating seamless collaboration and unlocking the full potential of research data.
Genomcore’s platform provides an intrinsic metadata system to ensure data is organized and structured effectively. This enables customized data models to be defined for each type of data, such as clinical and personal data, genomic data or the details of a laboratory experiment. Once the templates are defined, data can be recorded either manually through the web interface or programmatically using our API that is specifically designed for this purpose, which facilitates integrations with databases and other data sources and project partners.
The pseudo-anonymized data –being primary data and data resulting from the project analyses– is stored in the platform and is made accessible only to project partners via controlled user access. The platform provides a flexible database model to ensure segregation of information blocks -interoperable data structures- which allows defining granular permissions on data access on a user-level avoiding unforeseen usage or disclosure to non-authorized partners.

Genomcore’s platform also provides a collaborative, flexible and compliant hybrid computational infrastructure to meet the needs of the DIPCAN project, with a strong focus on compliance and collaborative frameworks by teams of experts from different backgrounds.
In traditional machine learning, models are often separated from the original data causing the model training process to be intermittent and non-iterative. This means that the models are trained once and the same trained model is used for all future predictions or decisions. However, this approach has some limitations. Our technological framework stores both data sources and computational resources in the same place, which allows adopting a continuous and iterative AI models training process. With this approach, models are constantly updated and trained with the latest data uploaded to the platform. This approach ensures that the models stay up-to-date and can learn and adapt over time, improving their overall performance.
One of the project’s goals is to change patient’s participation in their healthcare journey. Patients are expected to be active partners in the project, with rights and responsibilities, and through this empowerment increase their autonomy and involvement in the decision-making process with the ultimate objective of improving the cost-effectiveness and delivery of care. To do so, a mobile device application (App on both iOS and Android) has been specifically designed using Genomcore’s Frontdesk technology.
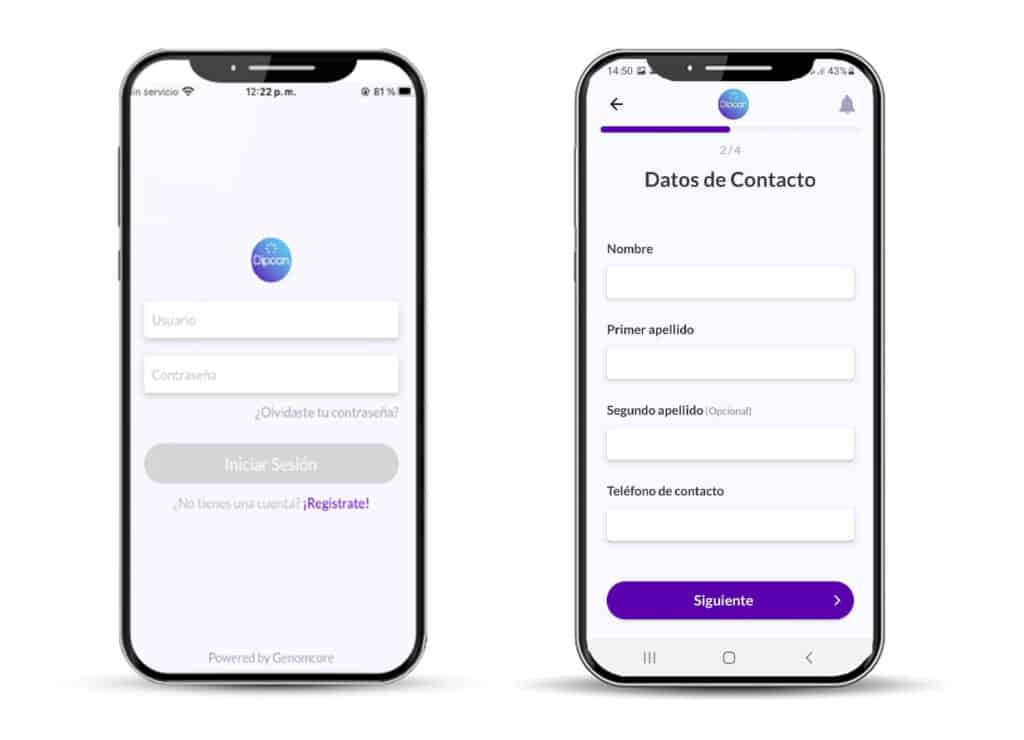
With the DIPCAN App, patients are proactively registered, healthcare professionals can easily retrieve patient’s health data through our questionnaire system, results reports are delivered in a user-friendly PDF format, and patient’s follow-up is carried out throughout the study with push notifications that facilitate direct communication between patients and healthcare professionals. As Genomcore Frontdesk is build onto Genomcore’s professional platform, the data retrieved is automatically saved and structured in the platform, which facilitates data collection, standardization and further exploitation using custom workflows or AI models.
“I believe the time has come to systematize the collection of this data in an intuitive, agile way that does not depend so much on the subjectivity of the doctor collecting it, and that specially will allow us to analyze and interpret this data in the future.”
Dr. Enrique Grande, the Principal Investigator and Head of the Medical Oncology Service and Clinical Research at the MD Anderson Cancer Center Madrid Foundation.
main goal
Multi-omics data has the potential to redefine the precision oncology paradigm. DIPCAN is a project where 7 leading institutions in Spain join forces to enhance the understanding of cancer in individual patients for earlier and more accurate diagnosis, targeted treatment, and a multidimensional analysis of tumor data. The AI models are expected not only to provide objective guidance and assistance in decision-making for individual clinical management of patients, but also to contribute to the development of national health strategies to manage these diseases in a more cost-effective manner. Collaboration among oncologists, surgeons, geneticists, bioinformaticians, pathologists, and molecular biologists is essential to advance the treatment of cancer.
Genomcore is proud to contribute with our expertise in multimodal data management and bioinformatic analysis to make personalized medicine a reality.
Discover the features used in this project


